CpG_Me:
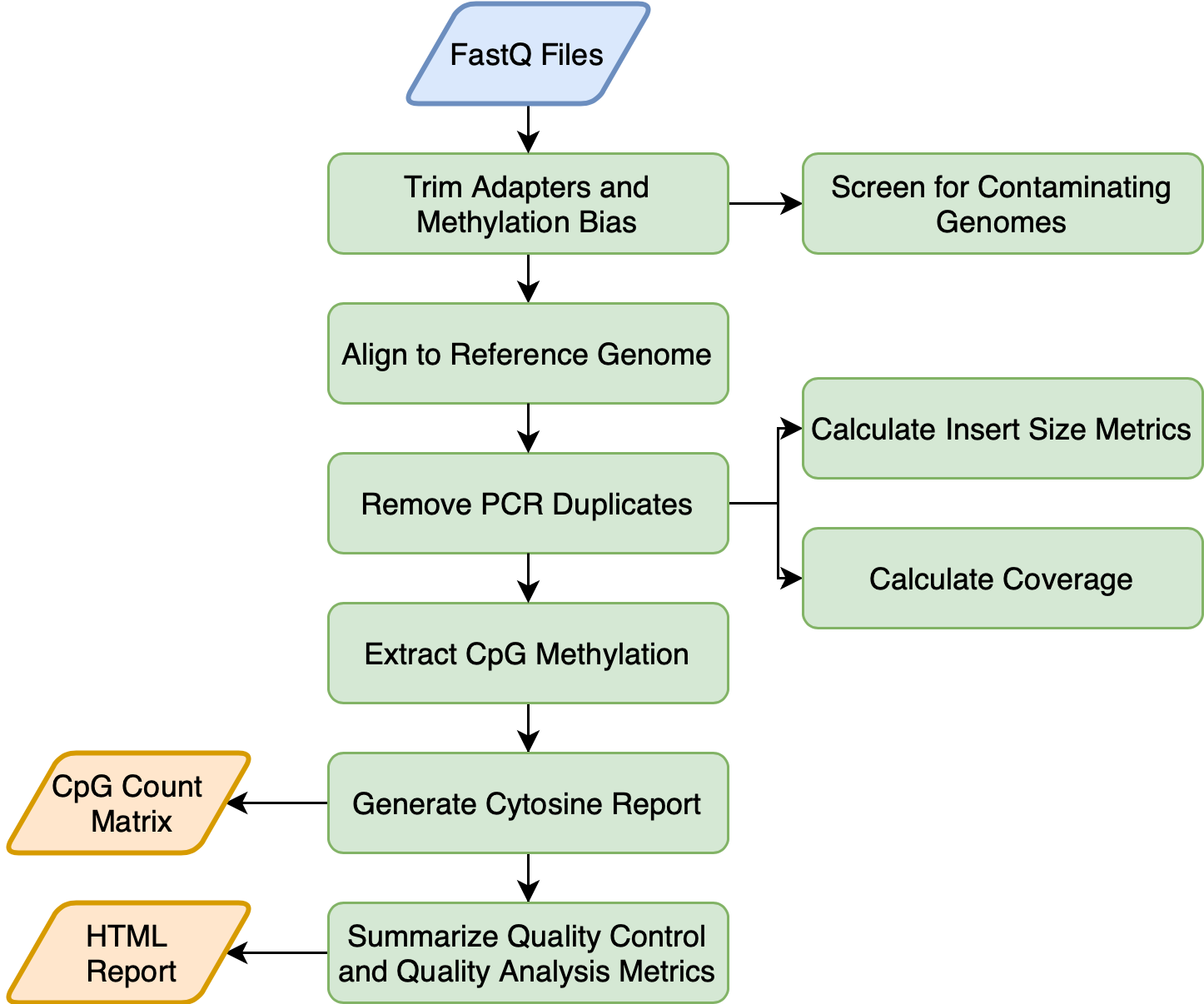
A whole-genome bisulfite sequencing (WGBS) pipeline for the analysis and QC of DNA methylation that goes from from raw reads to a CpG count matrix.
DMRichR:
An executable and package for the statistical analysis and visualization of differentially methylated regions (DMRs) from CpG count matrices (Bismark cytosine reports).


comethyl:
An R package for weighted region comethylation network analysis (Under development).

cnv-caller:
A copy number variation (CNV) caller that detects large-scale variation in low coverage aligned whole genome bisulfite sequencing (WGBS) data (Under development).

GEO Data:
Snord116-dependent diurnal rhythm of DNA methylation in mouse cortex
Integrative epigenomic analyses of early-life hypothalamic response to augmented maternal care
Role of DNMT3B in the Regulation of Early Neural and Neural Crest Specifiers
Large-Scale Methylation Domains Mark a Functional Subset of Neuronally Expressed Genes
Identifying Targets of MeCP2 During Neuronal Maturational Differentiation